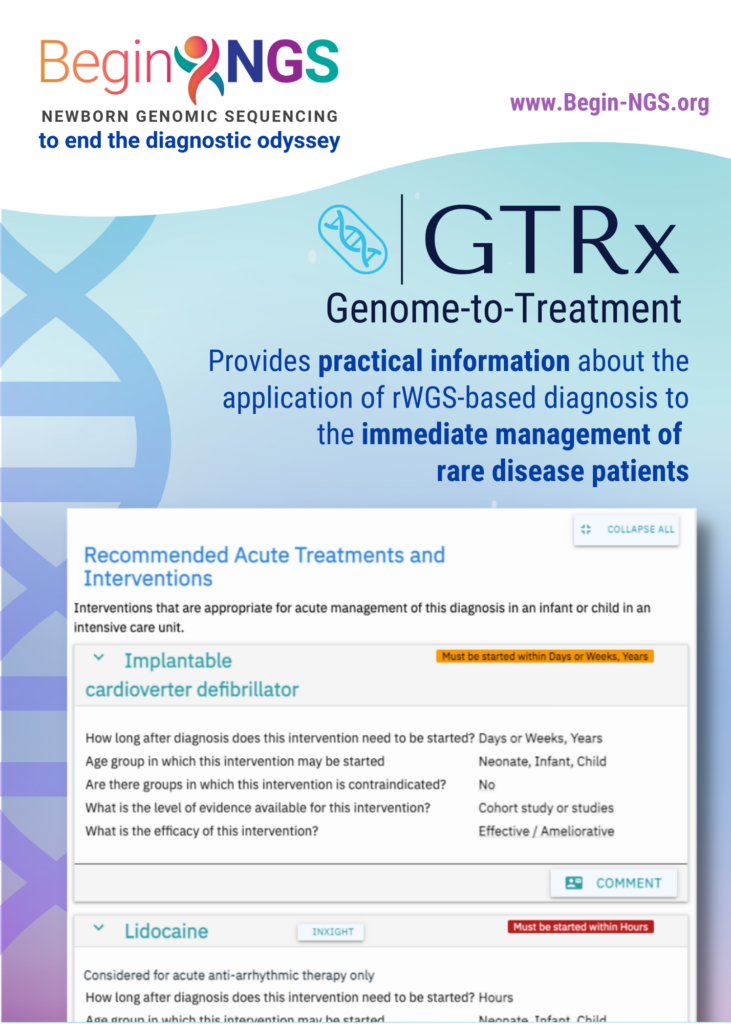
GTRx, a clinical decision support tool, can provide front-line clinicians with the information they need to initiate precision treatments for critically ill infants and children in intensive care
Rady Children’s Institute for Genomic Medicine (RCIGM®) today announced the publication of a study in Nature Communications describing and examining the performance of Genome-to-Treatment (GTRx™), an automated, virtual disease management system that integrates a rapid Whole Genome Sequencing (rWGS®) diagnosis completed in 13.5 hours with a custom lab information management system and analysis pipeline. Currently, GTRx is a prototype and encompasses about 500 genetic diseases that progress rapidly and have effective, available treatments.
The study was completed in collaboration with Alexion, AstraZeneca’s Rare Disease group, Genomenon, Fabric Genomics, Illumina, SL Data Strategies, Keck Graduate Institute, Rady Children’s Hospital–San Diego, and UC San Diego departments of Pediatrics and Neuroscience.
It currently takes an average of 5 years to diagnose a child with a rare disease1 and involves multiple specialists.
With that in mind, the authors developed a system designed for automated genetic disease diagnosis and acute management guidance. They began by reducing the turnaround time of rWGS to 13.5 hours, incorporating several innovations that enhanced scalability and reproducibility including automated interpretation and improving the ability to diagnose genetic diseases associated with most major classes of genomic variants. In addition, the team incorporated GTRx to provide immediate treatment guidelines for physicians to help them understand the identified genetic conditions and their available treatment options, which may include therapeutics, dietary changes, surgery, medical devices or other interventions.
GTRx is a critical piece of the recently announced diagnostic and precision medicine guidance tool called BeginNGS™ (pronounced “beginnings”) that would screen newborns for approximately 400 genetic diseases that have known treatment options using rapid Whole Genome Sequencing (rWGS®). BeginNGS, developed through a research collaboration with Alexion; Fabric Genomics; Genomenon; Illumina, Inc.; and TileDB, uses rWGS to diagnose and identify treatment options for genetic conditions before symptoms begin, an advancement over current pediatric uses of rWGS that focus mainly on children who are already critically ill. Once a diagnosis is made, BeginNGS uses GTRx to provide clinical decision support and help decrease variability or delayed implementation of optimal precision treatment following diagnosis of a rare genetic condition.
Alexion, one of the key collaborators on GTRx, integrated multiple online disease information resources and provided artificial intelligence tools to mine the published literature regarding available treatments for rare genetic diseases.
“By integrating tools to enable rapid diagnosis and deliver disease management guidance into an automated virtual system, GTRx is a remarkable example of how we can leverage digital technology to help doctors and patients get answers faster,” said Tom Defay, Alexion’s Deputy Head, Diagnostics Strategy and Development. “As a leader in rare diseases for 30 years, Alexion remains committed to helping shorten the diagnostic odyssey for patients, and we are proud of our collaboration to provide RCIGM with technical expertise and support for the development of this innovative technology.”
Another important partner in the development of GTRx, Illumina, contributed genome sequencing library preparation, sequencing, analysis, and data integration technologies that enabled the GTRx system.
“We at Illumina are very proud to play a role in this groundbreaking work,” said Ashley Van Zeeland, head of Illumina Open Innovation. “Our Customer Collaboration & Innovation team collaborated deeply with Dr. Kingsmore and team to develop solutions that set a new benchmark for enabling end-to-end precision care more quickly for these critically-ill babies.”
As part of the study, an expert clinical panel systematically reviewed published evidence about the efficacy of 9911 existing drugs, devices and dietary and surgical interventions for more than 500 severe, childhood genetic diseases. The clinical panel concluded that 1527 treatments had acceptable evidence of efficacy in 421 genetic diseases. These treatments, efficacy assessments, quality of supporting evidence, and indications and contraindications were integrated with 13 genetic disease information resources, such as GARD, Orphanet and OMIM, and appended to diagnostic reports.
“We developed GTRx to both increase the number of children who receive optimal, immediate treatment and to facilitate broader use of rWGS by physicians working in local hospitals who may not be as familiar with genetic diseases as sub- and super-specialists in regional, academic, tertiary or quaternary centers,” said Dr. Kingsmore, “Our goal is to upskill these physicians by providing patient diagnosis- and disease-specific information in real time.”
“Currently GTRx is available for research purposes,” added Dr. Kingsmore. “Upon validation of clinical utility, we will expand the system to additional genetic diseases and clinical use, as well as incorporating ongoing, expert, open community-based review.”
Co-authors include: Mallory J. Owen, Sebastien Lefebvre, Christian Hansen, Chris M. Kunard, David P. Dimmock, Laurie D. Smith, Gunter Scharer, Rebecca Mardach, Mary J. Willis, Annette Feigenbaum, Anna-Kaisa Niemi, Yan Ding, Luca Van DerKraan, Katarzyna Ellsworth, Lucia Guidugli, Bryan R. Lajoie, Timothy K. McPhail, Shyamal S. Mehtalia, Kevin K. Chau, Yong H. Kwon, Zhanyang Zhu, Sergey Batalov, Shimul Chowdhury, Seema Rego, James Perry, Mark Speziale, Mark Nespeca, Meredith S. Wright, Martin G. Reese, Francisco M. De LaVega, Joe Azure, Erwin Frise, Charlene Son Rigby, Sandy White, Charlotte A. Hobbs, Sheldon Gilmer, Gail Knight, Albert Oriol, Jerica Lenberg, Shareef A. Nahas, Kate Perofsky, Kyu Kim, Jeanne Carroll Nicole G. Coufal, Erica Sanford, Kristen Wigby, Jacqueline Weir, Vicki S. Thomson, Louise Fraser, Seka S. Lazare, Yoon H. Shin, Haiying Grunenwald, Richard Lee, David Jones, Duke Tran, Andrew Gross, Patrick Daigle, Anne Case, Marisa Lue, James A. Richardson, John Reynders, Thomas Defay, Kevin P. Hall, Narayanan Veeraraghavan, Stephen F. Kingsmore.
Funding for this research came, in part, from gifts made by Ernest and Evelyn Rady, grants from the National Institute of Child Health and Human Development (NICHD), National Human Genome Research Institute (NHGRI) and National Center for Advancing Translational Sciences (NCATS), and funds from Rady Children’s Hospital–San Diego.
Full study: An automated 13.5-hour system for scalable diagnosis and acute management guidance for genetic diseases.
Reference
1 Global Commission on Rare Disease [Internet]. [cited 2021 Dec 6]. Available from: https://www.globalrarediseasecommission.com/Report

